Computational and Mathematical Biology Centre (CMBC)
Aim and scope
Develop novel computational tools and mathematical models to address biological problems.
Enhance the fabric of research in computing through linkages with experiments and vice-versa.
To inculcate the application of emerging and relevant computational technologies for in-depth and advanced biological data analysis, structural mapping and for therapeutic intervention.
CMBC: A centre for discovery, innovation and translation
Major Program 1: Mathematical Modelling and Systems Biology
- Area of research:
(a) Mathematical modeling for understanding biological processes/ disease dynamics.
(b) Potential drug target discovery using network analysis and computational models.
Overview of the mathematical modelling and network analysis lab

- Research theme
Disease mechanism and potential targets through mathematical models and computational methods.
New mathematical and computational methods to study biological data.
konnect2prot (k2p) Click here

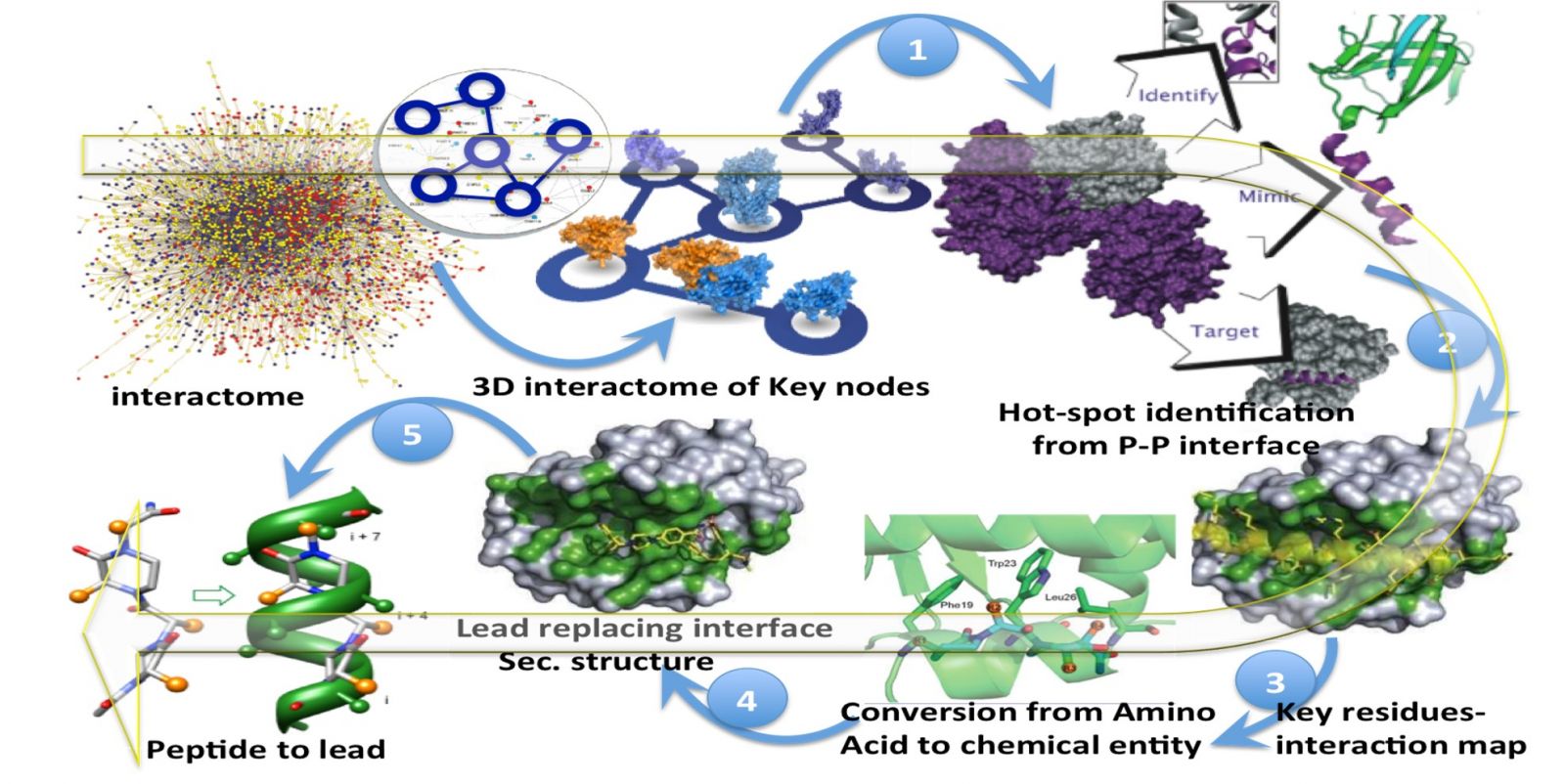
Major program 2: Computer-assisted Drug Discovery, Computational Biophysics and Structural Bioinformatics
- Area of research:
Virtual screening of synthetic compounds libraries for predicting potential hit molecules against specific drug targets.
Computer-aided drug discovery for infectious and metabolic diseases.
Computational simulations for molecular understanding of biological systems, mechanistic basis of structure-function correlation and their application to design therapeutics protein-protein interaction interfaces.
Computer-assisted drug discovery

Peptidomimetics: small molecules from active peptides
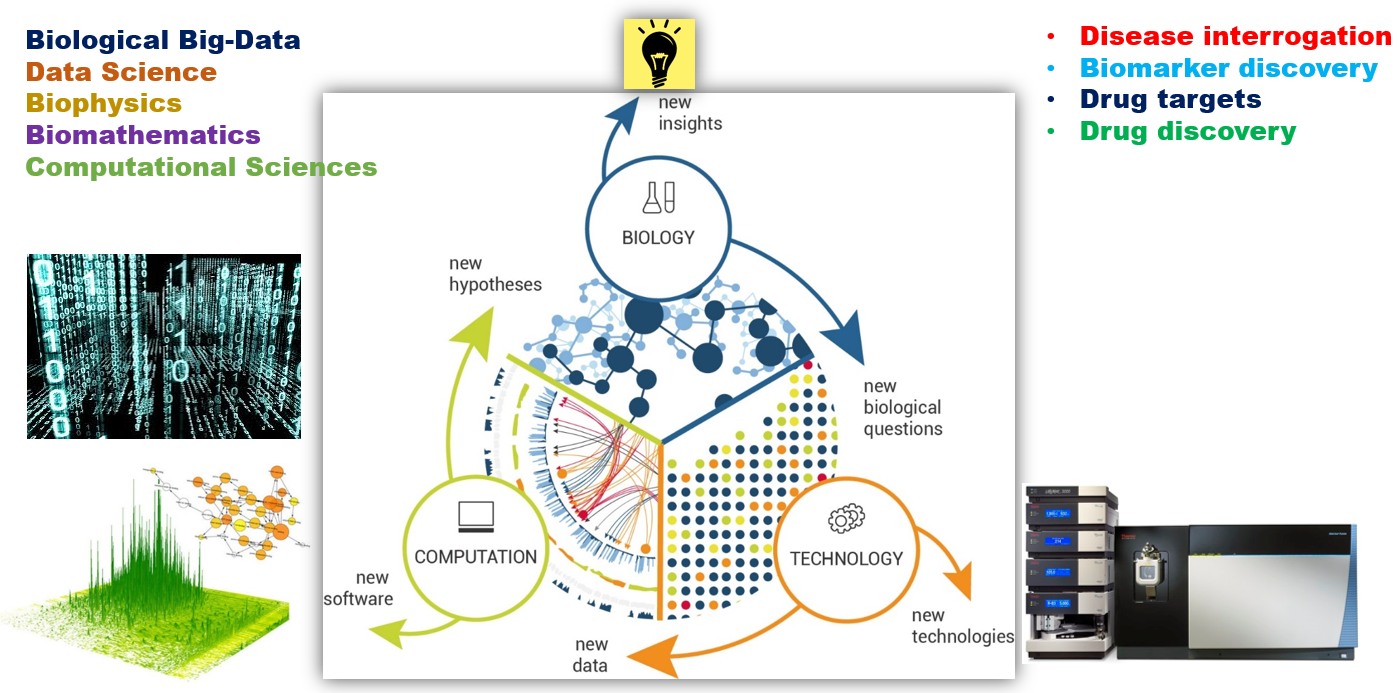
Major Program 3: Big data, Multi-Omics and Biomedical Informatics
- Area of research
Mass spectrometry based identification of proteins from biological samples to study disease progression in NAFLD.
Development of a universal proteogenomics workflow for integrating genomics/transcriptomics data with mass spectrometry proteomics data for studying liver proteoforms and studying their progression in disease progression.
Development of biomolecular knowledge resources platform from large scale multi-omics data for Human Liver proteoforms using proteogenomics to facilitate disease studies.
Development of a meta-resistome webserver for rapid and comprehensive mining of antimicrobial resistance genes (ARGs) from genomic and metagenomics data to infer the resistance potential of pathogen genomes/metagenomes.
Prioritization of Disease Proteins and Metabolites in NAFLD and NASH Using Big-Data Approaches
Many disease genes for NAFLD and NASH are known, but prioritization for drug target and biomarker use is necessary
This can be facilitated by integration of data on different biomolecules (genes, proteins, metabolites, PPIs, PTMs)
Big-data mining can reveal biomolecules that can be potential biomarkers or drug targets

Integrated analysis to find important proteins and metabolites

Meta-resistome web-server for mining AMR genes from NGS data

Name of faculty members and Scientists
DR. SAMRAT CHATTERJEE, PROFESSOR
DR. SHAILENDRA ASTHANA, PRINCIPAL SCIENTIST-II
DR. AMIT KUMAR YADAV, PRINCIPAL SCIENTIST-II
The centre/facility is open to providing services to academia and industry. For any queries, contact the following
Contact Details
Dr. Samrat Chatterjee
.png) samrat.chatterjee@thsti.res.in
samrat.chatterjee@thsti.res.in
.png) 0129- 2876479
0129- 2876479

